4. Multimodal Topic Models¶
Detailed description of all parameters and methods of BigARTM Python API classes can be found in Python Interface.
Now let’s move to more complex cases. In last section we mentioned the term modality. It’s something that corresponds to each token. We prefer to think about it as about the type of token, For instance, some tokens form the main text in the document, some form the title, some from names of the authors, some from tags etc.
In BigARTM each unique token has a modality. It is denoted as class_id (don’t confuse with the classes in the task of classification). You can specify the class_id of the token, or the library will set it to @default_class. This class id denotes the type of usual tokens, the type be default.
In the most cases you don’t need to use the modalities, but there’re some situations, when they are indispensable. For example, in the task of document classification. Strictly speaking, we will talk about it now.
You need to re-create all the data with considering the presence of the modalities. Your task is to create the file in the Vowpal Wabbit format, where each line is a document, and each document contains the tokens of two modalities - the usual tokens, and the tokens-labels of classes, the document belongs to.
Now follow again the instruction from the introduction part, dealing with your new Vowpal Wabbit file to achieve batches and the dictionary.
The next step is to explain your model the information about your modalities and the power in the model. Power of the modality is it’s coefficient 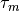 (we assume you know about it). The model uses by default only tokens of
(we assume you know about it). The model uses by default only tokens of @default_class modality and uses it with 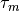 = 1.0. You need to specify other modalities and their weights in the constructor of the model, using following code, if you need to use these modalities:
= 1.0. You need to specify other modalities and their weights in the constructor of the model, using following code, if you need to use these modalities:
model = artm.ARTM(num_topics=20, class_ids={'@default_class': 1.0, '@labels_class': 5.0})
Well, we asked the model to take into consideration these two modalities, and the class labels will be more powerful in this model, than the tokens of the @default_class modality. Note, that if you had the tokens of another modality in your file, they wouldn’t be taken into consideration. Similarly, if you had specified in the constructor the modality, that doesn’t exist in the data, it will be skipped.
Of course, the class_ids field, as all other ones, can be reseted. You always can change the weights of the modalities:
model.class_ids = {'@default_class': 1.0, '@labels_class': 50.0}
# model.class_ids['@labels_class'] = 50.0 --- NO!!!
You need to update the weights directly in such way, don’t try to refer to the modality by the key directly: class_ids can be updated using Python dict, but it is not the dict.
The next launch of fit_offline() or fit_online() will take this new information into consideration.
Now we need to enable scores and regularizers in the model. This process was viewed earlier, excluding one case. All the scores of 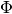 matrix (and perplexity) and
matrix (and perplexity) and 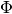 regularizers has fields to deal with modalities. Through these fields you can define the modalities to be deal with by score or regularizer, the other ones will be ignored (here’s the full similarity with
regularizers has fields to deal with modalities. Through these fields you can define the modalities to be deal with by score or regularizer, the other ones will be ignored (here’s the full similarity with topic_names field).
The modality field can be class_id or class_ids. The first one is the string containing the name of the modality to deal with, the second one is a list of strings.
Note
The missing value of class_id means class_id = @default_class, missing value of class_ids means usage of all existing modalities.
Let’s add the score of sparsity 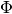 for the modality of class labels and regularizers of topic decorrelation for each modality, and start fitting:
for the modality of class labels and regularizers of topic decorrelation for each modality, and start fitting:
model.scores.add(artm.SparsityPhiScore(name='sparsity_phi_score',
class_id='@labels_class'))
model.regularizers.add(artm.DecorrelatorPhiRegularizer(name='decorrelator_phi_def',
class_ids=['@default_class']))
model.regularizers.add(artm.DecorrelatorPhiRegularizer(name='decorrelator_phi_lab',
class_ids=['@labels_class']))
model.fit_offline(batch_vectorizer=batch_vectorizer, num_collection_passes=10)
Well, we will leave you the rest of the work (tuning 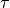 and
and 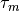 coefficients, looking at scores results etc.). In 5. Phi and Theta Extraction. Transform Method we will go to the usage of fitted ready model for the classification of test data.
coefficients, looking at scores results etc.). In 5. Phi and Theta Extraction. Transform Method we will go to the usage of fitted ready model for the classification of test data.